Siegel Lab
Infections with seemingly identical pathogens can lead to drastically different outcomes – as seen in the Corona pandemic. Such differences can be due to host specific factors or caused by cell-to-cell heterogeneity in pathogen populations.
The Siegel group aims to elucidate the mechanisms that control cell-to-cell heterogeneity in a population across different scales, i.e. how differences in genome sequence, chromatin organization and 3D genome architecture influence the expression of different surface antigens. A special focus of our work lies on understanding how ncRNA contribute to the organization of the 3D genome architecture and how RNA processing hotspots can serve as ‘post-transcriptional’ enhancers (Müller, Cosentino et al., Nature 2018, Faria, Luzak et al., Nature Microbiology 2021, Rabuffo et al., Nature Communications 2024).
Read more
Lab News
-
 Genomic Determinants of Antigen Expression Hierarchy in African Trypanosomes
Genomic Determinants of Antigen Expression Hierarchy in African TrypanosomesA highly sensitive single-cell RNA sequencing approach sheds light on the VSG-selection mechanisms in Trypanosoma brucei.
-
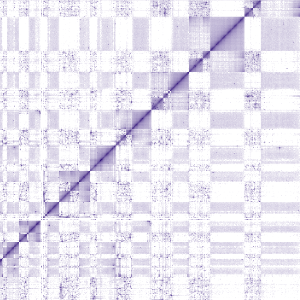
 Micro-C reveals T. brucei genome organisation at high resolution
Micro-C reveals T. brucei genome organisation at high resolutionInter-chromosomal transcription hubs shape the 3D genome architecture of African trypanosomes
-
 RNA processing and stability contribute to gene expression
RNA processing and stability contribute to gene expressionWe implemented SLAM-seq and TT-seq in African trypanosomes to study RNA maturation
-
 Tissue spaces are reservoirs of antigenic diversity for trypanosomes
Tissue spaces are reservoirs of antigenic diversity for trypanosomesSleeping sickness: pathogens play hide-and-seek. Great collaboration with the Mugnier lab