Genomic Determinants of Antigen Expression Hierarchy in African Trypanosomes
13 Mar 2025
A highly sensitive single-cell RNA sequencing approach sheds light on the VSG-selection mechanisms in Trypanosoma brucei.
13 Mar 2025
A highly sensitive single-cell RNA sequencing approach sheds light on the VSG-selection mechanisms in Trypanosoma brucei.
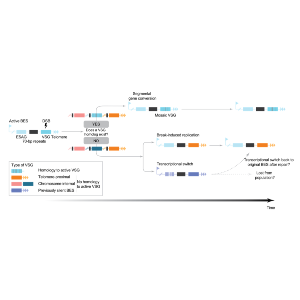
Our PhD student Zhibek Keneskhanova and our postdoc Dr. Raúl Cosentino, together with Dr. Kirsty McWilliam (former postdoc), in collaboration with the Colomé-Tatché and Mugnier labs, have developed a high-resolution, trypanosome-adapted single-cell RNA sequencing method (Spliced-Leader Smart‑seq3xpress) to capture VSG switching events in Trypanosoma brucei. Their work addresses the long-standing question of why certain VSGs are activated more frequently than others. By inducing a double-strand break in the active VSG gene, they demonstrate that both the availability of homologous repair templates and the genomic location of VSGs are critical determinants. These findings establish a new genomic framework for understanding antigen switching and immune evasion in African trypanosomes.
Keneskhanova Z#, McWilliam KR#, Cosentino RO#, Barcons-Simon A, Dobrynin A, Smith JE, Subota I, Mugnier MR, Colomé-Tatché M*, Siegel TN* (2025)
Genomic determinants of antigen expression hierarchy in African trypanosomes.
Nature https://doi.org/10.1038/s41586-025-08720-w